Next Generation Single-Cell Sequencing (NGS) is becoming a valuable standard method in life science research and personalized medicine.
Formalin-fixed paraffin-embedded (FFPE) tissue samples are a relevant source of material for this because of its wide availability and broad spectrum of sample types.
However, making high quality single cells from FFPE samples accessible for NGS is a challenge. Reflecting the tumor or tissue heterogeneity to uncover different genetic and phenotypic characteristics is a real bottleneck.

Tang et al. Cell Biosci. (2019) 9:53

FFPE 50 µm sections*
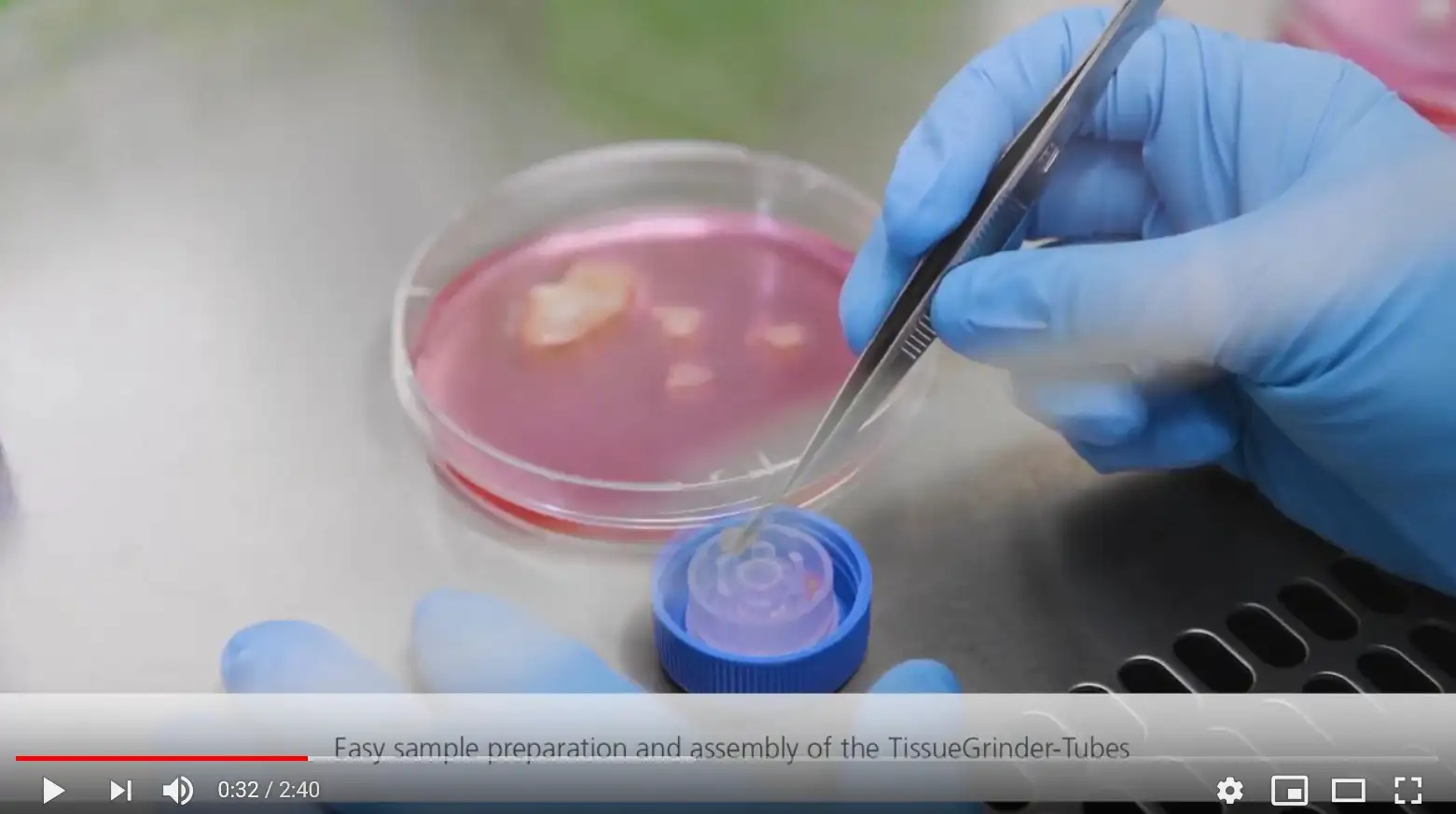
Minimized 20 min enzyme treatment
Fast but gentle mechanical dissociation with the TissueGrinder


Antibody labeling
Cytospins
FACS Isolation of pure tumor and stromal cell populations

Access to pure tumor and stromal cell DNA
Library preparation & detection of tumor specific characteristics
* TissueGrinder optimised FFPE protocol is based on the following study: https://link.springer.com/article/10.1186/s10020-019-0108-z
The 40x microscopy image show examples of how the single cells from FFPE samples generated with our protocol are suitable for immunofluorescent staining.
Keratin
Vimentin
DAPI




The single cells obtained with the TissueGrinder process are in a much better condition than those produced with a conventional enzymatic automation.

On the left, an example of single cells generated from FFPE samples with the TissueGrinder compared to a standard enzymatic dissociation on the right.
Above, an example of single cells generated from FFPE samples with the TissueGrinder compared to a standard enzymatic dissociation below.




Sounds too good to be true?
Then contact us to arrange a test run with your own samples!